Abstract
Introduction:
Infection remains an important cause of non-relapse mortality after allogeneic hematopoietic stem cell transplantation (allo-HSCT). Due to the impaired immune function of allo-HSCT patients, the clinical manifestations of pulmonary infection are diverse and the imaging characteristics are atypical. Meanwhile, pulmonary infections are often mixed and opportunistic pathogens are common. Therefore, diagnosis of these infections is difficult. Compared with the traditional culture method, metagenomic next-generation sequencing (mNGS) has the advantages of culture-independent, unbiased, short time-consuming and broad-spectrum detection. It is also able to detect rare and new pathogens. To further investigate the value of mNGS among allo-HSCT patients, we compared the results of mNGS of bronchoalveolar lavage (BAL) samples and microbiological culture among pulmonary infection patients after allo-HSCT.
Materials and Methods:
From May 28, 2018 to December 28, 2021, BAL samples were collected from 64 patients after allo-HSCT with pulmonary infection. mNGS and microbial culture of BAL were performed to evaluate the effectiveness of mNGS in the diagnosis of pulmonary infectious pathogens. Patients with disease relapse after allo-HSCT were excluded, and the follow-up was up to March 24, 2022.
Results:
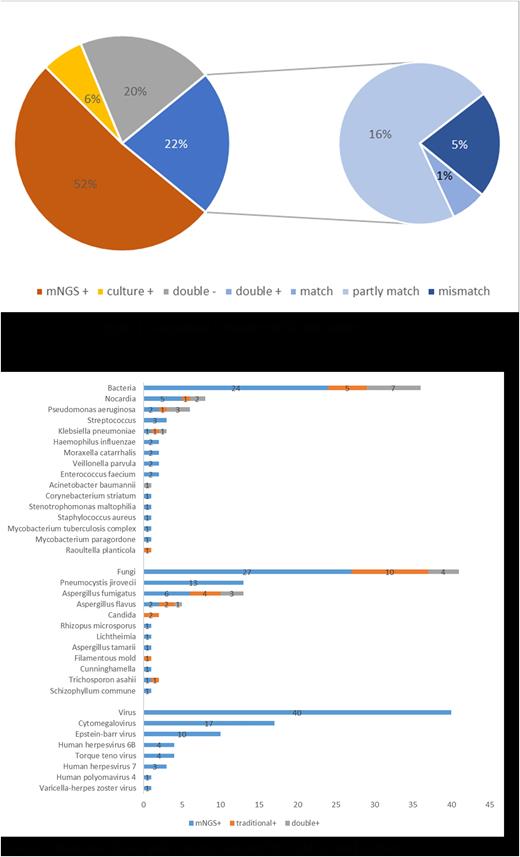
64 patients were enrolled in this study. The median age was 36 (17-67) years old, including 45 males (70.3%) and 19 females (29.7%), and the median time between transplantation and mNGS detection was 11.1 (0.7-91.2) months. Among 64 patients, the number of patients with only positive mNGS results was 33, 4 patients were only culture-positive, 13 patients were double negative, and 14 patients were double positive. For the double-positive group, 78.6% (11/14) of them showed completely or partly matched results. (Figure 1) In total, 47 patients (73.4%) were positive for mNGS and 18 patients (28.1%) were positive for culture, with the positive rate of mNGS higher than that of culture (p<0.01). Further analysis showed that the positive rate of fungi detected by mNGS was higher than that of culture (P = 0.043), and there was no significant difference between the positive rate of bacteria detected by mNGS and culture (P = 0.067), but the number of bacteria detected by mNGS (p=0.001) was higher than that of culture. Cytomegalovirus (CMV) (n=17) and Epstein Barr virus (EBV) (n=10) were the most commonly detected viruses in mNGS of BAL, and Pneumocystis yersini (n=13) was the most commonly detected fungus in BAL mNGS. (Figure 2) The number of Nocardia and Aspergillus fumigatus detected by BAL mNGS was higher than culture with no statistical significance. Previous the diagnosis of Pneumocystis carinii pneumonia (PCP) was based on the clinical manifestations, imaging examination, and Pneumocystis yersini detected by microscopic examination. Among 13 PCP patients, 12 patients (92.3%) had fever, cough,chest tightness,and shortness of breath, and 1 patient (7.7%) had no symptoms. 11 patients received G test, only one test result slightly exceeded the upper limit, and the other 10 tests were negative. 9 patients had typical pulmonary imaging changes of PJP, and 1 patient's CT finding of lung was consistent with the imaging findings of aspergillus pneumonia. The results showed that mNGS of BAL had advantage in the diagnosis of PCP after allo-HSCT.
Conclusion:
The study indicated that for patients with pulmonary infection after allo-HSCT, the positive rate of mNGS of BAL samples was higher than that of microbial culture especially for fungal infections. Furthermore, mNGS of BAL samples showed advantages over traditional methods in the detection of PCP after allo-HSCT.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal